Long-lasting foetal microchimerism in maternal brain is common, affects many brain regions
Thursday, 27 September 2012
First Evidence of Foetal DNA Persisting in Female Human Brain Tissue
Posted by ZenMaster at Thursday, September 27, 2012
Labels: brain, chimera, fetal, human, microchimerism, US 0 comments
Tuesday, 25 September 2012
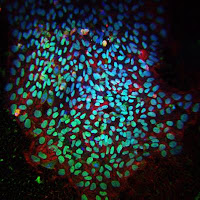
Making It Easier to Make Stem Cells
Sanford-Burnham researchers identified kinase inhibitors that lower the barrier to producing stem cells in the laboratory — cells important for disease research and drug development
 |
Induced pluripotent stem cells
generated
using a kinase inhibitor. Credit: Sanford-
Burnham
Medical Research Institute.
|
Posted by ZenMaster at Tuesday, September 25, 2012
Labels: California, iPS, research, stem cells 0 comments
What Can the Axolotl Teach Us About Tissue Regeneration In Humans?
Understanding how salamanders grow new limbs provides insights into the potential of human regenerative medicine
 |
Salk
researchers Gerald M. Pao, Wei
Zhu, and
Tony Hunter. Credit: the
Salk Institute for
Biological Studies.
|
Posted by ZenMaster at Tuesday, September 25, 2012
Labels: axolotl, California, differentiation, regenerative, research, transposon 0 comments
Friday, 21 September 2012
Discovery of Reprogramming Signature May Help Further Stem Cell-based Regenerative Medicine Research
Salk scientists show nine genes at heart of epigenetic changes in induced pluripotent stem cells
Posted by ZenMaster at Friday, September 21, 2012
Labels: California, epigenetic, iPS, research, stem cells, US 0 comments
Friday, 14 September 2012
New Efficiency to Stem Cell Reprogramming
Biologists reveal genes key to development of pluripotency, in single cells
Posted by ZenMaster at Friday, September 14, 2012
Labels: c-Myc, embryonic, iPS, Klf4, mouse, Oct4, reprogram, research, Sox2, stem cells, US 0 comments
Neural Stem Cells Regenerate Axons in Severe Spinal Cord Injury
New relay circuits, formed across sites of complete spinal transaction, result in functional recovery in rats
Posted by ZenMaster at Friday, September 14, 2012
Labels: California, human, neuron, rat, regenerative, research, stem cells 0 comments
Wednesday, 12 September 2012
Molecular Beacons Light Up Stem Cell Transformation
Molecular Beacons Light Up Stem Cell Transformation
Posted by ZenMaster at Wednesday, September 12, 2012
Labels: bone, differentiation, fat, reprogram, research, stem cells 0 comments
Tuesday, 11 September 2012
Neonatal Heart Stem Cells May Help Mend Kids' Broken Hearts
Cardiac stem cells from newborns show stronger regenerative ability than adult stem cells
Posted by ZenMaster at Tuesday, September 11, 2012
Labels: heart, human, research, stem cells 0 comments
Friday, 7 September 2012
How Many Genes in the Human Genome? Human Genome Far More Active than Thought
GENCODE Consortium discovers far more genes than previously thought
Posted by ZenMaster at Friday, September 07, 2012
Labels: chromosomes, DNA, gene expression, genome, human, lincRNAs, microRNA, sequence, UK 0 comments